Intrinsic Reaction Coordinate
Overview
The intrinsic reaction coordinate (IRC) maps a minimum energy path a reaction follows that connects the transition state to its reactants and products. IRC is run mainly to confirm whether the optimized transition state connects to the expected reactants and products. This helps in gaining insights into the reaction mechanism.
Check the Input and Visualizer section for the allowed input types and how to upload the files.
Modules Available
Presently, IRC can be run only with the DFT module. The task starts from an optimized transition state and descends along the paths in both the forward and backward directions. It follows the steepest descent method in which a Hessian is calculated mass-weighted step size.
DFT Module Input Fields
Upon selecting the DFT module, the following inputs must be provided:
Select the basis set family (e.g., Pople style, Ahlrichs)
Select the basis set (e.g., 6-31G, 6-31+G**)
Select the exchange functional category (e.g., Local and gradient method, Range-separated hybrid)
Choose an exchange-correlation functional (e.g., M06, B3LYP)
Total charge of the molecule (e.g., 0)
Spin multiplicity = 2S+1 (e.g., 1 for singlet)
Warning
IRC calculations have a high failure rate given the complex structure of the transition state.
Finally, click the Run IRC Calculation button to start the simulation.
Output Details
The reaction path visualization allows the user to explore the optimized geometries at different stages of the transition state search.
You can visualize the following structures from the dropdown menu:
Reactant Structure: Displays the initial geometry of the reactants.
Backward Trajectory: Shows the structures traced back from the transition state to the reactant.
Forward Trajectory: Displays the geometries along the path from the transition state to the product.
Full Trajectory: Combines both forward and backward paths for complete reaction insight.
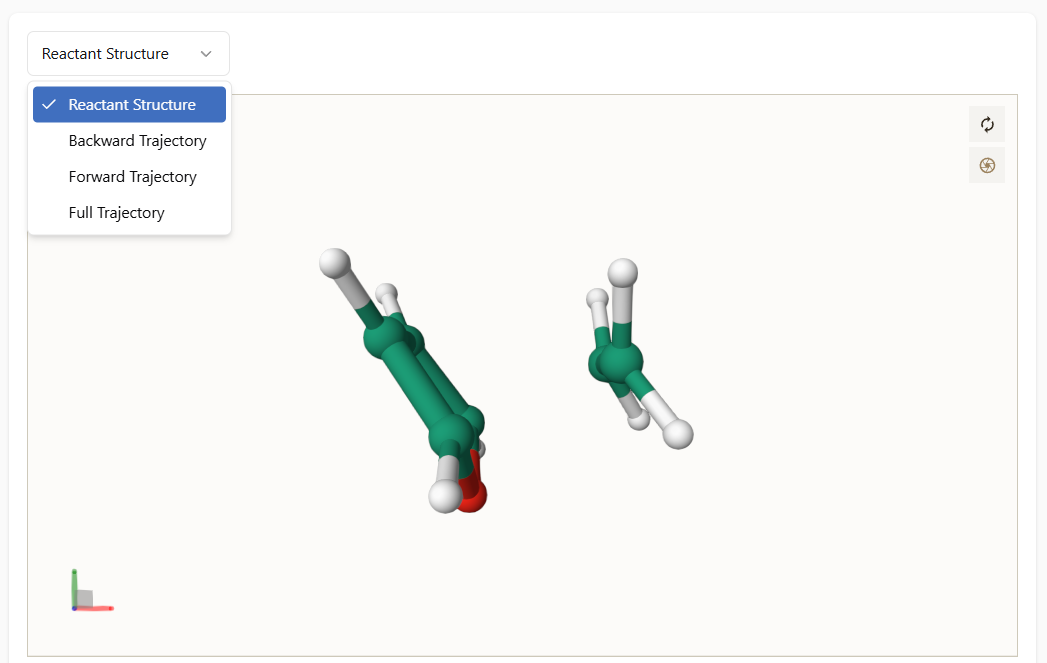
Users can download the trajectory files in XYZ format using the buttons located at the bottom-right corner of the visualizer.
Available downloads:
Forward Trajectory
Backward Trajectory
Full Trajectory
These files can be opened with any molecular viewer supporting .xyz format for further analysis.