Protein Preparation Task
Overview
Protein preparation is a crucial step in molecular docking workflows. Raw protein structures downloaded from public databases such as the Protein Data Bank (PDB) may contain missing atoms, multiple conformations, alternate locations, unwanted water molecules, and cofactors that are not relevant to docking. Preparing the protein structure ensures the active site is chemically accurate and geometrically optimized for meaningful interaction with ligands.
Why Protein Preparation is Important
Protein structures obtained from experimental methods like X-ray crystallography or NMR (as deposited in the PDB) are often incomplete or non-ideal for computational analysis.
Lack hydrogen atoms, which are essential for accurate docking and scoring.
Contain multiple conformations or alternate atom locations.
Include crystallographic water molecules that may interfere with docking.
Be in incorrect protonation states based on the docking pH.
Include bound ligands, ions, or cofactors that are irrelevant for your docking scenario.
Note
Without proper preparation, docking results may be unreliable, as the binding site may be misrepresented,leading to inaccurate predictions of ligand binding affinity and pose.
Preparation ensures a realistic, stable, and chemically correct receptor model.
Objectives
Clean the protein structure.
Remove irrelevant molecules (e.g., waters, heteroatoms, unnecessary chains).
Add hydrogens.
Convert to .pdbqt format for docking.
Component Description
The Protein Preparation interface includes:
- Input Protein Structure
Upload Structure File : Upload a .pdb file directly.
Use PDB ID : Provide a valid PDB ID (e.g., 1HSG) to fetch the structure from the PDB database.
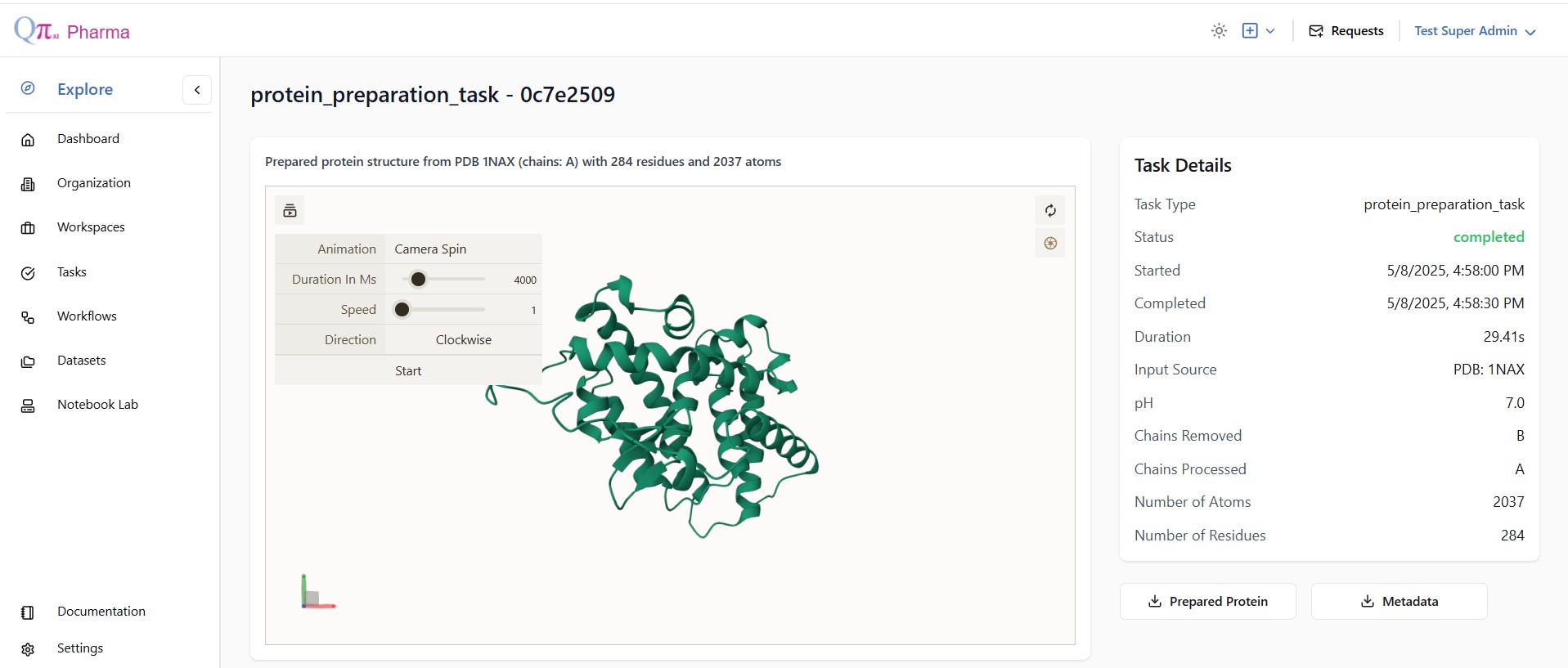
Protein Preparation
Chains to Remove : Specify unnecessary chains (e.g.,B,C,D) to exclude them from the final structure.
pH Value : Enter the pH at which hydrogen atoms should be optimized (default is 7.0).
Keep HETATM records : Check to retain heteroatoms (uncheck to remove).
Keep water molecules : Check to retain water (uncheck to remove).
Run Protein Preparation
Click the Run Protein Preparation button to process the input structure and generate a .pdbqt file.
How to Use the Interface
+-----------------------------+
| 1. Input Protein Structure |
+-----------------------------+
| |
v v
+------------------+ +---------------------------+
| Upload .pdb File | | Use PDB ID (e.g. 1HSG) |
+------------------+ +---------------------------+
\ /
\ /
v v
+----------------------+
| 2. Protein Preparation |
+----------------------+
|
v
+---------------------------------------------+
| Preparation Options |
| |
| Chains to remove: B, C, D |
| Target pH value: 7.0 |
| Keep HETATM records: [✔] or [ ] |
| Keep water molecules: [✔] or [ ] |
+---------------------------------------------+
Output Details
Once the protein is prepared user can visualise it and can download the .pdbqt file that can further used in the molecular docking.
Note
No missing atoms or residues are fixed.
This task is optimized for speed and basic cleanup.