Input and Visualizer
Overview
Our software provides an intuitive and interactive 3D molecular visualizer, designed to support a seamless user experience. It allows users to input molecular structures through multiple commonly used formats and immediately visualize them in a 3D environment.
Supported Input File Formats
The input file in the following file formats can be uploaded:
XYZ
SDF
PDB
PDBQT
To visualize a molecule:
Click on the Upload File(s) button.
Drag and drop one or more supported files.
The 3D molecular structure will be rendered instantly in the visualizer.
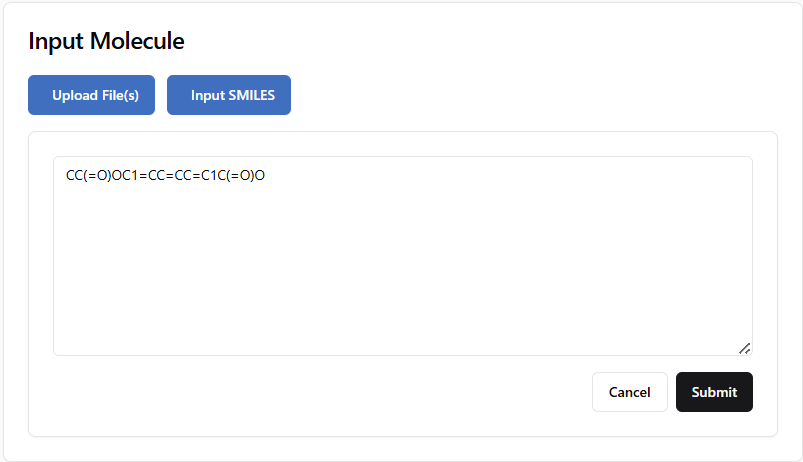
SMILES Input
In addition to file uploads, users can also input molecular structures using SMILES (Simplified Molecular Input Line Entry System) notation:
Enter a valid SMILES string into the input field.
Click the Submit button.
The corresponding molecule will appear in the 3D visualizer.
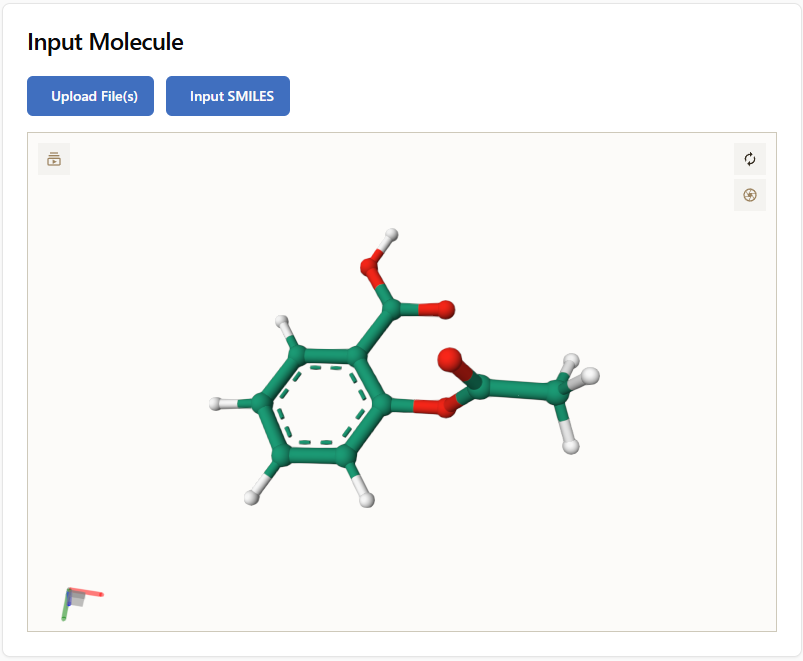
Example
Atom Index
To enhance the user experience, the visualizer displays atom indices on the 3D structure. This feature allows users to easily identify and reference specific atoms within the molecule. The atom indices are shown as labels next to each atom, providing a clear and immediate reference for users. This functionality is particularly useful for tasks such as: Constrained Optimization Details, Potential Energy Surface Scans.
This figure illustrates the atom indices displayed on a 3D structure:
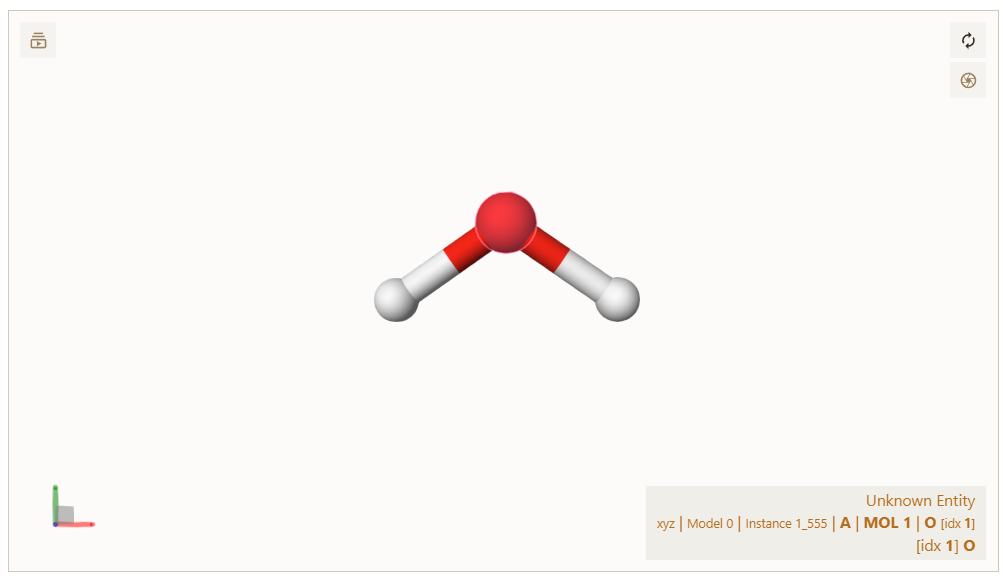
Atom indices displayed in the visualizer, here for a water molecule. O is the oxygen atom, and idx 1 is the index of the oxygen atom.